Visualizing the Palmer Penguins Dataset
In this post I will outline how to construct a data visualisation of the Palmer Penguins data set.
Downloading the Data
This first step might seem a little obvious, but the first thing we have to do before constructing any sort of data visualisation of a data set, we must first download and read in the data. To do so, I will be used the Pandas package and running the following lines of code:
import pandas as pd
import tabulate
url = "https://raw.githubusercontent.com/PhilChodrow/PIC16B/master/datasets/palmer_penguins.csv"
penguins = pd.read_csv(url)
Now, we may check the penguins dataset we have read into Python. This step may not be wholly necessary, however it is good practice to view the data set to make sure it is read in correctly.
penguins.head()
| studyName | Sample Number | Species | Region | Island | Stage | Individual ID | Clutch Completion | Date Egg | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | Comments | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | PAL0708 | 1 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A1 | Yes | 11/11/07 | 39.1 | 18.7 | 181.0 | 3750.0 | MALE | NaN | NaN | Not enough blood for isotopes. |
| 1 | PAL0708 | 2 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A2 | Yes | 11/11/07 | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE | 8.94956 | -24.69454 | NaN |
| 2 | PAL0708 | 3 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A1 | Yes | 11/16/07 | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE | 8.36821 | -25.33302 | NaN |
| 3 | PAL0708 | 4 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A2 | Yes | 11/16/07 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | Adult not sampled. |
| 4 | PAL0708 | 5 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N3A1 | Yes | 11/16/07 | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE | 8.76651 | -25.32426 | NaN |
Cleaning the Dataset
Once we have read in the data set, we must clean it before we start constructing a data visualization. This might involve removing rows with NaN values, dropping unecessary or constant columns, and improving readability.
Specifically for the Palmer Penguins data set, I will be dropping the following columns: studyName, Sample Number, Individual ID, Clutch Completion, Date Egg, and Comments. The reason for doing so is because even though useful information could be potentially derived from these variables, I personally do not find this information as useful.
columns_to_drop = ['studyName', 'Sample Number', 'Individual ID', 'Clutch Completion', 'Date Egg', 'Comments']
# creating a new sub-data set with the above columns dropped
pen = penguins.drop(columns_to_drop, axis=1)
# viewing the new data set
pen.head()
| Species | Region | Island | Stage | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | 39.1 | 18.7 | 181.0 | 3750.0 | MALE | NaN | NaN |
| 1 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE | 8.94956 | -24.69454 |
| 2 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE | 8.36821 | -25.33302 |
| 3 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE | 8.76651 | -25.32426 |
Now that we have dropped some of the unecessary columns, we now need to search for any constant columns (i.e., columns with only one unique value) that do not add any useful information to the data set
columns_to_drop = []
for column in pen.columns:
# If the number of unique elements in a column
# is equal to 1, drop the column
if pen[column].nunique() == 1:
columns_to_drop.append(column)
# Viewing the constant columns
columns_to_drop
['Region', 'Stage']
Since Region and Stage are constant columns, we may also remove those columns from the data set as follows.
pen = pen.drop(columns_to_drop, axis=1)
# Viewing the data set
pen.head()
| Species | Island | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | MALE | NaN | NaN |
| 1 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE | 8.94956 | -24.69454 |
| 2 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE | 8.36821 | -25.33302 |
| 3 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE | 8.76651 | -25.32426 |
The next step in the data cleaning process is to remove any NaN values from the data set. Luckily for us, there is a very handy pandas function i.e., dropna that let’s us do this:
pen = pen.dropna()
pen.head()
| Species | Island | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE | 8.94956 | -24.69454 |
| 2 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE | 8.36821 | -25.33302 |
| 4 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE | 8.76651 | -25.32426 |
| 5 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | MALE | 8.66496 | -25.29805 |
| 6 | Adelie Penguin (Pygoscelis adeliae) | Torgersen | 38.9 | 17.8 | 181.0 | 3625.0 | FEMALE | 9.18718 | -25.21799 |
Now, it would be a good idea to reset the index of the data set as we have dropped a good number of rows
pen = pen.reset_index(drop=True)
pen.tail() # checking the last few rows of the data set
| Species | Island | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | |
|---|---|---|---|---|---|---|---|---|---|
| 320 | Gentoo penguin (Pygoscelis papua) | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | FEMALE | 7.99184 | -26.20538 |
| 321 | Gentoo penguin (Pygoscelis papua) | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | FEMALE | 8.41151 | -26.13832 |
| 322 | Gentoo penguin (Pygoscelis papua) | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | MALE | 8.30166 | -26.04117 |
| 323 | Gentoo penguin (Pygoscelis papua) | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | FEMALE | 8.24246 | -26.11969 |
| 324 | Gentoo penguin (Pygoscelis papua) | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | MALE | 8.36390 | -26.15531 |
Now that we have dropped all the columns and rows we needed to and reset the index, the last step in our data cleaning process is to improve upon the readability of our dataset. Specifically, I want to improve on the readability of the columns with string data: Species, Island, and Sex columns. Consider the unique elements of the Species column
pen['Species'].unique() #extracting the unique elements of the species column
array(['Adelie Penguin (Pygoscelis adeliae)',
'Chinstrap penguin (Pygoscelis antarctica)',
'Gentoo penguin (Pygoscelis papua)'], dtype=object)
Notice that the individual strings are rather long. Instead, we could replace these strings with something a bit more readable. For instance, I would replace 'Adelie Penguin (Pygoscelis adeliae)' with 'Adelie', and so on.
# We will replace the appropriate Species names with 'Adelie', 'Chinstrap', and 'Gentoo'
pen['Species'] = pen['Species'].replace('Adelie Penguin (Pygoscelis adeliae)', 'Adelie')
pen['Species'] = pen['Species'].replace('Chinstrap penguin (Pygoscelis antarctica)', 'Chinstrap')
pen['Species'] = pen['Species'].replace('Gentoo penguin (Pygoscelis papua)', 'Gentoo')
pen['Species'].unique() # checking the unique species
array(['Adelie', 'Chinstrap', 'Gentoo'], dtype=object)
Similar to what we have done with the Species column, we will also investigate the unique elements of the Island and Sex columns.
# Checking the unqiue elements of the Island column
pen['Island'].unique()
array(['Torgersen', 'Biscoe', 'Dream'], dtype=object)
The readability of the unique elements in the Island column is pretty good as it is, so we do not need to make any changes!
# Checking the unique elements of the Sex column
pen['Sex'].unique()
array(['FEMALE', 'MALE', '.'], dtype=object)
Here we have the standard 'FEMALE' and 'MALE' sex elements, but we also have a third element '.'. For the sake of simplicity, I will be replacing the '.' element with NaN and removing the appropriate rows from the data set
# Removing all the '.'from the Sex column for simplicity
pen['Sex'] = pen['Sex'].replace('.', float('NaN'))
pen = pen.dropna() # removing the NaN values
pen # Checking the dataset
| Species | Island | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE | 8.94956 | -24.69454 |
| 1 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE | 8.36821 | -25.33302 |
| 2 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE | 8.76651 | -25.32426 |
| 3 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | MALE | 8.66496 | -25.29805 |
| 4 | Adelie | Torgersen | 38.9 | 17.8 | 181.0 | 3625.0 | FEMALE | 9.18718 | -25.21799 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 320 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | FEMALE | 7.99184 | -26.20538 |
| 321 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | FEMALE | 8.41151 | -26.13832 |
| 322 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | MALE | 8.30166 | -26.04117 |
| 323 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | FEMALE | 8.24246 | -26.11969 |
| 324 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | MALE | 8.36390 | -26.15531 |
324 rows × 9 columns
We have now successfully cleaned our data set!
Constructing the Visualization
The first step before constructing the visualization is to import the relevant visualization packages: Matplotlib, Seaborn, etc.
import seaborn as sns
from matplotlib import pyplot as plt
The type of visualization I am interested in constructing involves a few of the characteristic features of penguins: Culmen Length (mm), Culmen Depth (mm), Flipper Length (mm), Body Mass (g), Delta 15 N (o/oo), Delta 13 C (o/oo). I will have to pick two of these features for my visualization. To do so, I will construct a pairplot of the data set and choose the two features with the most interesting set of data clusters:
sns.set_theme(style="ticks")
sns.pairplot(pen, hue="Species")
plt.suptitle('Palmer Penguins Pairplot', x=0.5, y=1.0)
plt.savefig('palmer-penguins-pairplot.png')

The pairplot is one useful way of visualizaing the Palmer Penguins data set as it shows data relationships between the various features or columns. However, while we retain the information on the Species of penguins, we also do end up leaving out the Island and Sex information.
After studying the pairplot, I thought that the plot between Culmen Length (mm) and Culmen Depth (mm) was pretty cool as it has three distinct clusters, one corresponding to each species. As such, I have chosen those two features for my visualization.
plt.figure(figsize=(20, 10))
fig = sns.relplot(
data=pen, x="Culmen Length (mm)", y="Culmen Depth (mm)",
col="Island", hue="Species", style="Sex",
kind="scatter"
)
plt.suptitle('Classification of Palmer Penguins by Island, Sex, and Species', x=0.5,y=1.05)
plt.savefig('palmer-penguins-classification.png')
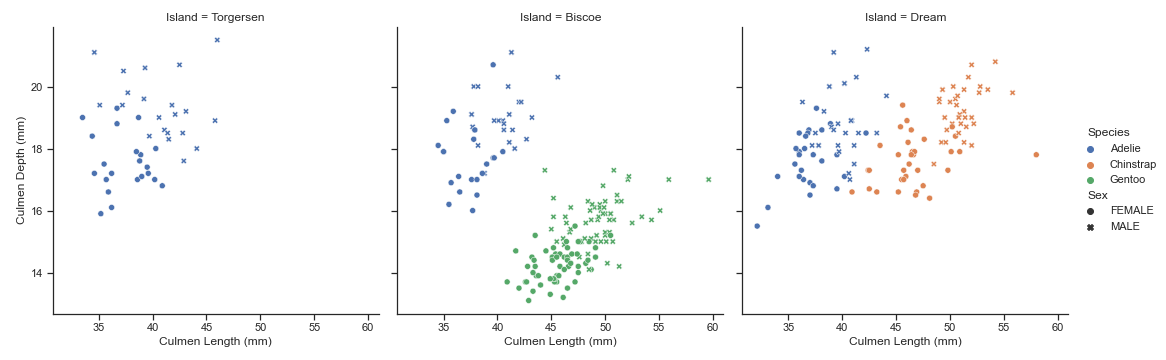
As such, with the above final visualization, I have three plots (one corresponding to each Island) depicting the relationship between Culmen Length (mm) and Culmen Depth (mm). Additionally, I have also changed up the markers to differentiate between the Sex of the penguins.
We are able to extract some interesting insights from this particular visualization; for instance, we note that only Adelie penguins are found in the Torgersen Island, Adelie and Gentoo are found in Biscoe Island, and Adelie and Chinstrap are found on Dream Island. We are also able to extract useful insights about the various Species of penguins from the data clusters!
The best part about this visualization is that we could switch up the x and y axes with other features and extract useful insights from those clusters as well!
Something I found really troublesome in this project was the data exploration and cleaning phase. However, though it was a tad bit taxing, I found that it really helped me in the end as I was able to generate my data visualization super easily. To be sure, a lesson learned the hard way is to always properly clean (and profile) the data set before attempting anything else!
Disclaimer
This blog post was created for my PIC16B class at UCLA (Previously titled: Blog Post 0 - Visualizing the Palmer Penguins Dataset).
